Developing Multi-Step Workflows
Last updated on 2025-01-14 | Edit this page
Estimated time: 0 minutes
Overview
Questions
- How can we expand to a multi-step workflow?
- What is iterative workflow development?
- How to use workflows as dependency graphs?
- How to use sketches for workflow design?
Objectives
- Explain that a workflow is a dependency graph
- Use cwlviewer online
- Generate Graphviz diagram using cwltool
- Exercise with the printout of a simple workflow; draw arrows on code; hand draw a graph on another sheet of paper
- Recognise that workflow development can be iterative i.e. that it doesn’t have to happen all at once
- Understand the flow of data between tools
Multi-Step Workflow
In the previous episode we worked through a single step workflow,
carrying out quality control check on RNA reads of the fruitfly genome.
In this episode the workflow is extended with an equivalent reverse RNA
reads and the next two steps of the RNA-sequencing analysis, trimming
the reads and aligning the trimmed reads, are added. We will be using
the cutadapt and
STAR tools for these
tasks.
To make a multi-step workflow that can carry all this analysis out,
we add more entries to the steps field.
Naming steps
Note that when the quality_control step is duplicated
the two steps are named quality_control_forward and
quality_control_reverse, to distinguish the separate
forward and reverse RNA reads. Likewise, the
rna_reads_fruitfly input becomes
rna_reads_fruitfly_forward, and an
rna_reads_fruitfly_reverse input is added.
YAML
cwlVersion: v1.2
class: Workflow
inputs:
rna_reads_fruitfly_forward:
type: File
format: http://edamontology.org/format_1930 # FASTQ
rna_reads_fruitfly_reverse:
type: File
format: http://edamontology.org/format_1930 # FASTQ
ref_fruitfly_genome: Directory
fruitfly_gene_model: File
steps:
quality_control_forward:
run: bio-cwl-tools/fastqc/fastqc_2.cwl
in:
reads_file: rna_reads_fruitfly_forward
out: [html_file]
quality_control_reverse:
run: bio-cwl-tools/fastqc/fastqc_2.cwl
in:
reads_file: rna_reads_fruitfly_reverse
out: [html_file]
trim_low_quality_bases:
run: bio-cwl-tools/cutadapt/cutadapt-paired.cwl
in:
reads_1: rna_reads_fruitfly_forward
reads_2: rna_reads_fruitfly_reverse
minimum_length: { default: 20 }
quality_cutoff: { default: 20 }
out: [ trimmed_reads_1, trimmed_reads_2, report ]
mapping_reads:
requirements:
ResourceRequirement:
ramMin: 5120
run: bio-cwl-tools/STAR/STAR-Align.cwl
in:
RunThreadN: {default: 4}
GenomeDir: ref_fruitfly_genome
ForwardReads: trim_low_quality_bases/trimmed_reads_1
ReverseReads: trim_low_quality_bases/trimmed_reads_2
OutSAMtype: {default: BAM}
SortedByCoordinate: {default: true}
OutSAMunmapped: {default: Within}
Overhang: { default: 36 } # the length of the reads - 1
Gtf: fruitfly_gene_model
out: [alignment]
index_alignment:
run: bio-cwl-tools/samtools/samtools_index.cwl
in:
bam_sorted: mapping_reads/alignment
out: [bam_sorted_indexed]
outputs:
quality_report_forward:
type: File
outputSource: quality_control_forward/html_file
quality_report_reverse:
type: File
outputSource: quality_control_reverse/html_file
bam_sorted_indexed:
type: File
outputSource: index_alignment/bam_sorted_indexedThe workflow file shows the first 5 steps of the RNA-seq analysis:
quality_control_reverse,
quality_control_forward,
trim_low_quality_bases, mapping_reads, and
index_alignment.
The index_alignment step uses the alignment
output of the mapping_reads step. You do this by
referencing the output of the mapping_reads step in the
in field of the index_alignment step. This is
similar to referencing the outputs of the different steps in the
outputs section.
Default values
The mapping_reads step needs some extra information
beyond the inputs from the other steps, which is done by providing
default values. If you want, you can read the
bio-cwl-tools/STAR/STAR-Align.cwl file to see how these
extra inputs are transformed into command line options to the
STAR program. This information is provided in the
in field.
Specifying resources
To run the tool better, it needs more RAM than the default. So there
is a requirements entry inside the
mapping_reads step definition with a
ResourceRequirement to allocate a minimum of 5120 MiB (5
GiB) of RAM.
The newly added mapping_reads step also need an input
not provided by any of our other steps, therefore an additional
workflow-level input is added: a directory that contains the reference
genome necessary for the mapping.
This ref_fruitfly_genome is added in the
inputs field of the workflow and in the YAML input file,
workflow_input_2.yml.
Validating the workflow
Is this a valid workflow? 🌶
Use cwltool to validate the workflow
A warning is thrown after we validate this workflow.
WARNING Workflow checker warning:
rna_seq_workflow_2.cwl:51:11: Source 'alignment' of type ["File", {"type": "array", "items":
"File"}] may be incompatible
rna_seq_workflow_2.cwl:56:7: with sink 'bam_sorted' of type "File"Should we be concerned about this warning 🌶🌶🌶
Not all warnings are bad.
Should we be concerned about this warning? If not, why not?
In this case, the outputs of the mapping step may instead either be just one file, OR an array of files.
It is important that the invoke the mapping step in such
a way that only one file is output, as a single file is the requirement
of the index_alignment step
Running the new workflow
The workflow definition is complete and we now only need to write the YAML input file.
YAML
rna_reads_fruitfly_forward:
class: File
location: rnaseq/GSM461177_1_subsampled.fastqsanger
format: http://edamontology.org/format_1930 # FASTQ
rna_reads_fruitfly_reverse:
class: File
location: rnaseq/GSM461177_2_subsampled.fastqsanger
format: http://edamontology.org/format_1930 # FASTQ
ref_fruitfly_genome:
class: Directory
location: rnaseq/dm6-STAR-index
fruitfly_gene_model:
class: File
location: rnaseq/Drosophila_melanogaster.BDGP6.87.gtfWe have finished the workflow definition and the input file and now can run the workflow.
Challenge: Draw the workflow 🌶
Draw the connecting arrows in the following graph of our workflow. Also, provide the outputs/inputs of the different steps. You can use for example Paint or print out the graph.
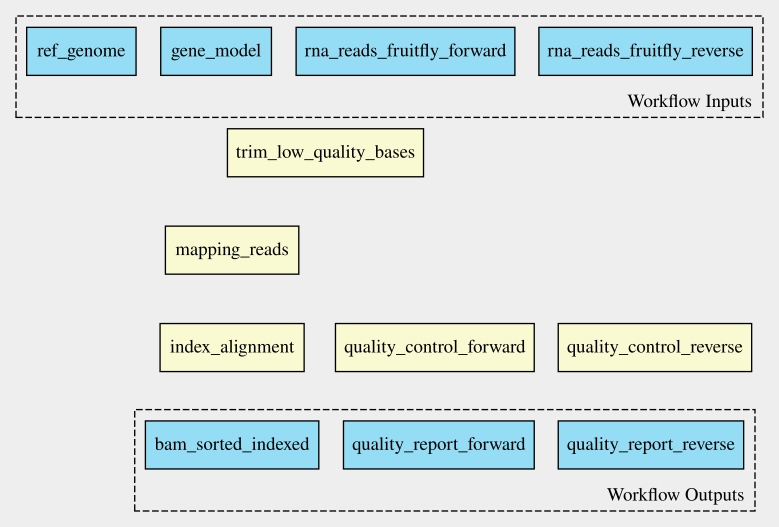
To find out how the inputs and the steps are connected to each other,
look at the in field of the different steps.
Iterative working
Working on a workflow is often not something that happens all at once. Sometimes you already have a shell script ready that can be converted to a CWL workflow. Other times it is similar to this tutorial, you start with a single-step workflow and extend it to a multi-step workflow. This is all iterative working, a continuous work in progress.
Visualising a workflow
To visualise a workflow, a graph can be used. This can be done before
a CWL script is written to visualise how the different steps connect to
each other. It is also possible to make a graph after the CWL script has
been written. This graph can be generated using online tools or the
built-in function in cwltool. When a graph is generated, it
can be used to visualise the steps taken and could make it easier to
explain a workflow to other researchers.
A CWL workflow is a directed acyclic graph (DAG). This means that:
- The workflow has a certain direction, from workflow inputs to step inputs, from step outputs to other step inputs, and from step outputs to workflow outputs and
- The workflow definition has no cycles.
CWL workflow as a dependency graph
By being a ‘DAG’, a CWL workflow is a dependency graph. Each input for a step in the workflow depends on either a workflow-level input or the presence of a particular output from another step.
From CWL script to graph
In this example the workflow is already made, so the graph can be
generated using cwlviewer
online or using cwltool. First, let’s have a look at cwlviewer. To use this tool, the
workflow has to be put in a GitHub, GitLab or Git repository. To view
the graph of the workflow enter the URL and click
Parse Workflow.
Push your workflow to GitHub 🌶
Add your workflow to a git commit and then push that commit to github.com
Now it’s time to view your workflow!
View your workflow in the cwl viewer 🌶
Paste the workflow url into the form on view.commonwl.org
Your workflow url will be something like
https://github.com/alexiswl/cwl-novice-tutorial/blob/main/rna_seq_workflow_2.cwl.
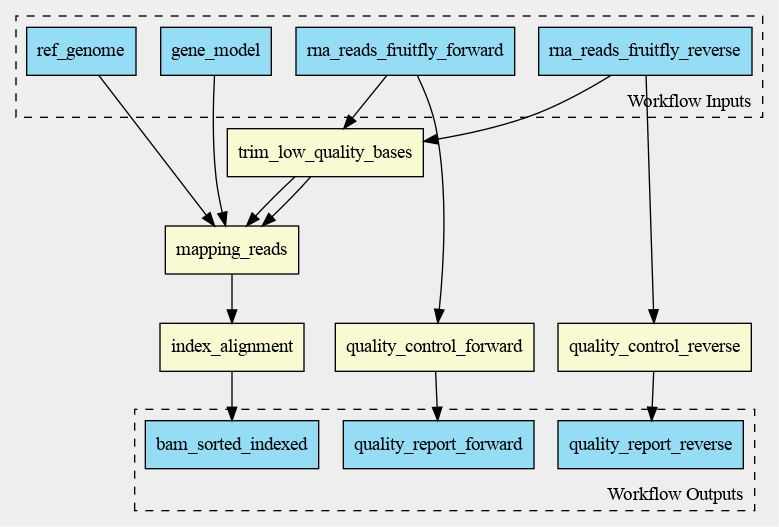
The cwlviewer displays the workflow as a graph, starting with the input. Then the different steps are shown, each with their input(s) and output(s). The steps are linked to each other using arrows accompanied by the input of the next step. The graph ends with the workflow outputs.
The graph of the RNA-seq workflow looks a follows:

Generating graphs locally
It is also possible to generate the graph in the command line.
cwltool has a function that makes a graph. The
--print-dot option will print a file suitable for Graphviz
dot program. This is the command to generate a Scalable
Vector Graphic (SVG) file:
The resulting SVG file displays the same graph as the one in the
cwlviewer. The SVG file can be opened in any web browser and in Inkscape, for example. Or opened with
code workflow_graph_2.svg from the terminal.
Windows Only: View images from the CLI with wslview
Windows users can run wslview workflow_graph_2.svg in
their terminal to view the graph in the default web browser.
Visualisation in VSCode
Benten is an extension in Visual Studio Code (VSCode) that among other things visualises a workflow in a graph. When Benten is installed in VSCode, the tool can be used to visualise the workflow. In the top-right corner of the VSCode window the CWL viewer can be opened, see the screenshot below.
.png)
In VSCode/Benten the inputs are shown in green, the steps in blue and
the outputs in yellow. This graph looks a little bit different from the
graph made with cwlviewer or cwltool. The graph by
VSCode/Benten doesn’t show the output-input names between the different
steps.
.png)
Key Points
- A multi-step workflow has multiple entries under the
stepssection - Workflow development can be an iterative process
- A CWL workflow can be represented as a dependency graph, either to explain your workflow or as a planning tool
